Planctomycetes
Planctomycetes are a phylum of widely distributed Bacteria, occurring in both aquatic and terrestrial habitats.[1] They play a considerable role in the global carbon and nitrogen cycles, with many species of this phylum capable of performing anaerobic ammonium oxidation, also known as anammox.[1][2] Many Planctomycetes also often associate with other organisms such as macroalgae and marine sponges.[3]
| Planctomycetes | |
|---|---|
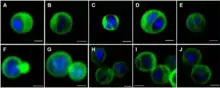 | |
| Confocal laser scanning micrographs of Gemmata obscuriglobus budding cells stained with DAPI and DiOC6 . | |
| Scientific classification | |
| Domain: | Bacteria |
| Superphylum: | |
| Phylum: | Planctomycetes Garrity & Holt 2001 |
| Classes | |
| |
| Synonyms | |
| |
Planctomycetes are included in the PVC superphylum along with Verrucomicrobia, Chlamydiae, Lentisphaerae, Kiritimatiellaeota, and Candidatus Omnitrophica.[4][5] The Planctomycete phylum is composed of two classes, Planctomycetia and Phycisphaerae. First described in 1924, Planctomycetes were first identified as eukaryotes and were only later described as bacteria in 1972.[1] Early examination of Planctomycetes suggested a cell plan differing considerably from all other known bacteria, although they are now confirmed as gram-negative bacteria having many unique characteristics.
The morphology of Planctomycetes are often small spherical cells, however there is a large amount of variation.[6] Planctomycetes also display distinct reproductive habits with many species dividing by budding, in contrast to all other free-living bacteria which divide by binary fission.[1][7][8]
There is also growing interest in Planctomycetes and biotechnology, mainly as a source of bioactive molecules.[9] Some Planctomycetes were recently described as human pathogens.[3]
The species Gemmata obscuriglobus has been identified as being unique among Planctomycetes.[10][11]
Structure and Morphology
Cell shape and appendages
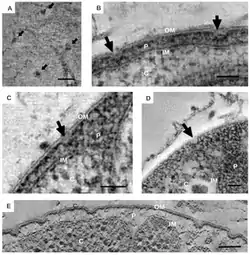
The distinct morphological characteristics of Planctomycetes have been discussed extensively in scientific research.[2] The common morphology is often spherical cells roughly 2 μm in diameter, as observed in the species Aquisphaera giovannonii. However, the diversity in cell shape often varies greatly in Planctomycetes. Ovoid and pear-shaped cells have been described in some species, and often occur in rosettes of 3-10 cells.[6] Gemmata obscuriglobus is a well studied species of Planctomycete with spherical cells. In contrast, the species Planctopirus limnophila has ovoid cells.[10]
Many Planctomycetes display structures and appendages on the outer surface of the cell. Flagella, common in most bacteria, have been observed in P. limnophila.[1][6][12] Many Planctomycetes also have a holdfast, or stalk, which attaches the cell to a surface or substrate.[1][12] Although, some species such as Isosphaera pallida lack a holdfast.[1]
Unique appendages known as crateriform structures have been observed in some Planctomycetes,[1][6][12] and seem to be specific to species of the class Planctomycetia.[8] The outer surface of P. limnophila displays both large and small crateriform structures. The large crateriform structures often cover the cell surface, while the small crateriform structures are often only at the end of the cell. Light microscopy demonstrated fibers of both stalk and pili type in P. limnophila and G. obscuriglobus. The pili fibers in P. limnophila and G. obscuriglobus were often associated with large crateriform structures. In contrast, the stalk fibers were associated with small crateriform structures.[12]
Cell wall composition
Early examination of Planctomycetes suggested that their cell plan differed considerably from both gram-positive and gram-negative bacteria. Until recently, Planctomycetes were thought to lack peptidoglycan in their cell walls, and were instead suggested to have proteinaceous cell walls. Peptidoglycan is an essential polymer of glycans, present in all free-living bacteria, and its rigidity helps maintain integrity of the cell. Peptidoglycan is also essential during cell division. Recently, Planctomycetes such as G. obscuriglobus, among many other species, were found to have peptidoglycan in their cell walls.[1][12]
Internal cell composition
Planctomycetes were once thought to display distinct compartmentalization within the cytosol.[1][12] Three-dimensional electron tomography reconstruction of G. obscuriglobus displayed varying interpretations of this suggested compartmentalization.[11] The cytosol was suggested to be separated into compartments, both the paryphoplasm and pirellulosome, by an intracytoplasmic membrane. This interpretation has since been demonstrated to be incorrect. In fact, the intracytoplasmic membrane is well known to be the cytoplasmic membrane which displays unique invaginations, giving the appearance of compartmentalization within the cytosol.[1][11][12] Planctomycetes therefore display the two compartments typical of Gram-negative bacteria, the cytoplasm and periplasm.
The excess membrane observed in G. obscuriglobus triples the surface area of the cell relative to its volume, which is suggested to be associated with sterol synthesis.[11]
Pigments
Many Planctomycetes display pink or orange coloring, suggested to result from the production of carotenoid pigments. Carotenoids are produced by plants and fungi, and by some heterotrophic bacteria to protect against oxidative stress. Three different carotenoid pigments have been identified in two different strains of Planctomycetes.[13]
In marine environments, Planctomycetes are often suspended in the water column or present as biofilms on the surface of macroalgae, and are often exposed to harmful ultraviolet radiation. Research suggests that more highly pigmented species of Planctomycetes are more resistant to ultraviolet radiation, although this is not yet well understood.[14]
Unique Characterisitcs of anammox cells
Anammox Planctomycetes form the order Brocadiales.[15] The cells of anammox bacteria are often coccoid with a diameter of roughly 0.8 μm.[16] The cell of anammox bacteria is suggested to contain three compartments, each surrounded by a membrane. The outer membrane encloses the cell and the protoplasm and the innermost membrane surrounds the amoxosome, the central structure of anammox bacteria. Anammox membrane composition resembles all other living organisms, as they are composed of glycerolipid.[17] Species of anammox bacteria contain a distinct compartment known as the anammoxosome.[12]
Life History and Reproduction
Growth
Planctomycetes grow slowly, when compared to other bacteria,[1][5][16][18] often forming rosette structures of 3-5 cells.[1][18] The species P. limnophila is suggested to be relatively fast growing,[1][19] with a doubling time of roughly 6-14 days. In contrast, some other Planctomycetes have doubling times of around 30 days.[19] The high abundance of Planctomycetes in many ecosystems is surprising, given their slow growth rates.[16][5]
_and_in_current_PVC_species_(Right).jpg.webp)
Life cycle
Planctomycetes often perform a lifestyle switch between both a sessile stalked stage and a free swimming stage.[18] P. limnophila performs a lifestyle switch that is often associated with cell division. The sessile mother cell produces a free swimming daughter cell. The daughter cell must then attach to a surface before starting the cycle over again. However, not all Planctomycetes have a motile stage and some research has suggested that the lifestyle switch observed in many species is not common among all Planctomycetes.[1]
.jpg.webp)
Reproduction
The current understanding of bacterial cell division is based on model organisms such as Escherichia coli.[10] The dominant form of reproduction observed in almost all bacteria is cell division by binary fission, which involves the synthesis of both peptidoglycans and proteins known as FtsZ.[10][20] In contrast, many Planctomycetes divide by budding.[1][7][8]
FtsZ proteins are suggested to be similar in structure to that of tubulin, the protein present in eukaryotes,[21] and is essential for septal formation during cell division.[1][2] The lack of FtsZ proteins is often lethal.[1] Peptidoglycan also play a considerable role in cell division by binary fission.[20]
Planctomycetes are one of the only known phyla lacking FtsZ proteins.[1][20][21] Chlamydiales, also a member of the PVC superphylum, also lack FtsZ.[21] Although Planctomycetes lack FtsZ, two distinct modes of cell division have been observed.[1] Most Planctomycetes divide by binary fission, including species of the class Phycisphaerae. In contrast, species of the class Planctomycetia divide by budding.[1][7][8]
The mechanisms involved in budding have been described extensively for yeast cells. However, bacterial budding observed in Planctomycetes is still poorly understood.[10] Budding has been observed in both radial symmetric cells, such as P. limnophila, and axial symmetric cells.[8] During cell division in P. limnophila, the daughter cells originate from the region opposite to the pole with the holdfast or stalk.
Considerable diversity has been observed in cell division among Planctomycetes.[7][8] During cell division in Fuerstia marisgermanicae, a tubular structure is connected from the bud to the mother cell.[1][15] The species Kolteria novifilia forms a distinct clade of Planctomycetes, and is the only known species to divide by lateral budding at the middle of the cell. Lastly, members of the clade Saltatorellus are capable of switching between both binary fission and budding.[7][8]
Genetic Characteristics
Genetic signatures
Although the Planctomycetes are renowned for their unusual cellular characteristics, their distinctness from all other bacteria is additionally supported by the shared presence of two conserved signature indels (CSIs).[22] These CSIs demarcate the group from neighboring phyla within the PVC group.[23] An additional CSI has been found that is shared by all Planctomycetes species, with the exception of Kuenenia stuttgartiensis, which is in line with the observation that K. stuttgartiensis forms a deep branch within the phylum.
A conserved signature indel has also been found to be shared by the entire PVC group, including Planctomycetes.[22][23] Planctomycetes also harbors an important conserved signature protein that has been characterized to play an important housekeeping function that is exclusive to members belonging to the PVC clade.[24]
General characteristics
The genome size of Planctomycetes has been observed as roughly 7,145,576 bases in the species Rhodopirellula baltica and is one of the largest prokaryotic circular genome sequences which has had its genome fully sequenced. This is mostly made up of multiple copies of the same sequence of around one thousand pairs of genes. This takes up around 25.4% of the genome sequence which is created by the process of extensive genome duplication.[2] This may be a way for the organism to adapt for mutations, as if one part were to be disabled, a different part would be copied altogether. The PCR primer which is used often mismatches with the genes creating difficulty when sequencing the genome.[4] When comparing under a microscope, A defining characteristic for these types of Planctomycetes is that a single, unlinked rRNA operon can be identified near the origin. There is a large inversion at position of 87,500 to 431,000 of the genome sequencing that twists the genome into a skewed shape. The changes of genetic material is through internal chromosomal inversion and not through lateral gene transfer. This creates a way of diversification in the planctomycetes variants as multiple transposes genes in these regions have reverse orientation that transfers to rearrangements.
Research concluded that Planctomycetes thrive in high concentrated nitrate environments. This could be the fact that the genes that are expressed which is required for heterotactic acid fermentation are present. Enzymes that are key in this, lactate dehydrogenase, play a key role in this process. The genetic process also has UV protection response. The genes that are responsible for these are recA, lexA, uvrA, uvrB, uvrC and one photolyase gene that is expressed when the environment offers excessive UV radiation stress. Other stress responses include the decomposition of hydrogen peroxide and oxidation. Sulfatase genes are also expressed by Planctomycetes as they are metabolically important on the survival of Planctomycetes. The Pirellula sp. strain 1 genome incorporates 110 genes which contributes to encoding proteins that produces sulfatase. This genome is identified to be more than two times the amount present in other prokaryotic species’ genome, indicated that these strive in high sulfate concentrated environments. In comparison with a different species of prokaryotic, the Pseudomonas aeruginosa, it can be found that there is only a maximum of 6 sulfatases and the genes that express this protein are contained as 2 to 5 pairs and is usually clustered in 22 groups. The functionality of these genes are meant to hydrolyze sulfate ester bonds which occur between glucose in heteropolysaccharides and metabolize the broken down sulfate for consumption.[2]
Phylogeny
Originally classified as a eukaryote due to morphology, the advent of genetic sequencing allowed researchers to agree that planctomycetes belong to the domain bacteria.[1] Within that domain planctomycetes are classified as their own phylum, however, other researchers have argued it could also be categorized as part of a larger superphylum entitled PVC which would encompass the phyla Verrucomicrobia, Chlamydiae and Lentisphaerae, as well as Poribacteria and the candidate phylum Candidatus Omnitrophica.[4] Within this superphylum its members have been found to be closely related through the creation of 16S rRNA trees. Both Planctomycetes and Chlamydiae encode proteins for nucleotide transporters, and Verrucomicrobia has also been found to have features common among eukaryotic cells. Thus it has been suggested that a common ancestor of this superphylum may have been the start of the eukaryotic lineage.[4] While this is one possible explanation, due to the fact that PVC is not the start of the bacterial tree,[25] it is more likely that the existence of eukaryotic traits and genes is explained through lateral gene transfer, and not a more recent eukaryotic ancestor.[4]
The phylogeny based on the work of the All-Species Living Tree Project.[26]
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Taxonomy
Within the Planctomycete phylum, there are two classes - Planctomycetia and Phycisphaerae. To date only 35 species within the Planctomycete phylum have been identified and 28 of them belong to the family Planctomycetaceae. There is also the candidate family Brocadiaceae which encompasses the species of Planctomycetes able to perform anammox. However, due to the distinct ability to perform anammox this family may be represented as its own class in the future.[1]
The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LSPN)[27] and the National Center for Biotechnology Information (NCBI).[28]
- Phylum "Planctomycetes" Garrity & Holt 2001 emend. Ward 2011 [Planctomycetaeota Oren et al. 2015]
- Class Phycisphaerae Fukunaga et al. 2010
- Order Tepidisphaerales Kovaleva et al. 2015
- Family Tepidisphaeraceae Kovaleva et al. 2015
- Genus Tepidisphaera Kovaleva et al. 2015
- Species Tepidisphaera mucosa Kovaleva et al. 2015
- Genus Tepidisphaera Kovaleva et al. 2015
- Family Tepidisphaeraceae Kovaleva et al. 2015
- Order Phycisphaerales Fukunaga et al. 2010
- Family Phycisphaeraceae Fukunaga et al. 2010
- Genus Algisphaera Yoon, Jang & Kasai 2014
- Species Algisphaera agarilytica Yoon, Jang & Kasai 2014
- Genus Phycisphaera Fukunaga et al. 2010
- Species Phycisphaera mikurensis Fukunaga et al. 2010
- Genus Algisphaera Yoon, Jang & Kasai 2014
- Family Phycisphaeraceae Fukunaga et al. 2010
- Order Tepidisphaerales Kovaleva et al. 2015
- Class Planctomycetacia Cavalier-Smith 2002
- Species ?"Candidatus Nostocoida limicola III" ♠ Liu et al. 2001
- Species ?"Candidatus Nostocoida acidiphila" ♠ Kulichevskaya et al. 2012
- Order "Brocadiales" Jetten et al. 2011
- Family "Brocadiaceae" Jetten et al. 2011
- Genus "Candidatus Anammoxoglobus" Kartal et al. 2006
- Species "Candidatus Anammoxoglobus propionicus" Kartal et al. 2006
- Genus "Candidatus Brocadia" Jetten et al. 2001
- Species "Candidatus B. anammoxidans" Jetten et al. 2001
- Species "Candidatus B. brasiliensis" Araujo et al. 2011
- Species "Candidatus B. caroliniensis" Park et al. 2017
- Species "Candidatus B. fulgida" Kartal et al. 2004
- Species "Candidatus B. sinica" Hu et al. 2010
- Genus "Candidatus Jettenia" Quan et al. 2008
- Species "Candidatus Jettenia asiatica" Quan et al. 2008
- Species "Candidatus Jettenia caeni" Ali et al. 2015
- Species "Candidatus Jettenia moscovienalis" Nikolaev et al. 2015
- Genus "Candidatus Kuenenia" Schmid et al. 2000
- Species "Candidatus Kuenenia stuttgartiensis" Schmid et al. 2000
- Genus "Candidatus Scalindua" Schmid et al. 2003
- Species "Candidatus S. arabica" Woebken et al. 2008
- Species "Candidatus S. marina" Van de Vossenberg et al. 2007
- Species "Candidatus S. profunda" Van De Vossenberg et al. 2008
- Species "Candidatus S. richardsii" Fuchsman et al. 2012
- Species "Candidatus S. wagneri" Schmid et al. 2003
- Species "Candidatus S. sorokinii" Kuypers et al. 2003
- Species Candidatus S. brodae Schmid et al. 2003
- Genus "Candidatus Anammoxoglobus" Kartal et al. 2006
- Family "Brocadiaceae" Jetten et al. 2011
- Order Planctomycetales Schlesner & Stackebrandt 1987 emend. Ward 2011
- Family Planctomycetaceae Schlesner & Stackebrandt 1987 emend. Scheuner et al. 2014 [Isosphaeraceae Kulichevskaya et al. 2015]
- Genus "Bythopirellula" ♠ Storesund & Ovreas 2013
- Species "Bythopirellula goksoyri" ♠ Storesund & Ovreas 2013
- Genus "Candidatus Anammoximicrobium" ♠ Khramenkov et al. 2013
- Species "Candidatus Anammoximicrobium moscowii" ♠ Khramenkov et al. 2013
- Genus Aquisphaera Bondoso et al. 2011
- Species Aquisphaera giovannonii Bondoso et al. 2011
- Genus Blastopirellula Schlesner et al. 2004
- Genus Gemmata Franzmann and Skerman 1985
- Species Gemmata obscuriglobus Franzmann & Skerman 1985
- Genus Gimesia Scheuner et al. 2015 [Planctomyces maris (ex Bauld & Staley 1976) Bauld & Staley 1980]
- Species Gimesia maris Scheuner et al. 2015 [Planctomyces maris (ex Bauld & Staley 1976) Bauld & Staley 1980]
- Genus Isosphaera Giovannoni et al. 1995
- Species Isosphaera pallida (ex Woronichin 1927) Giovannoni et al. 1995 nom. rev. ["Isocystis pallida" Woronichin 1927; "Torulopsidosira pallida" (Woronichin 1927) Geitler 1963]
- Genus Paludisphaera Kulichevskaya et al. 2015
- Species Paludisphaera borealis Kulichevskaya et al. 2015
- Genus Pirellula Schlesner & Hirsch 1987 emend. Schlesner et al. 2004 [Pirella Schlesner & Hirsch 1984 non Bainier 1883]
- Species Pirellula staleyi Schlesner & Hirsch 1987 [Pirella staleyi Schlesner & Hirsch 1984 ill.]
- Genus Planctomicrobium Kulichevskaya et al. 2015
- Species Planctomicrobium piriforme Kulichevskaya et al. 2015
- Genus Planctomyces Gimesi 1924
- Species P. bekefii ♪ Gimesi 1924 (type sp.) ["Blastocaulis sphaerica" Henrici & Johnson 1935; "Acinothrix globulifera" Novácek 1938; "Planctomyces subulatus" Wawrik 1952; "Planctomyces crassus" Hortobágyi 1965]
- Species P. guttaeformis ♪ (ex Hortobágyi 1965) Starr and Schmidt 1984
- Species P. stranskae ♪ (ex Wawrik 1952) Starr and Schmidt 1984
- Genus Planctopirus Scheuner et al. 2015
- Species Planctopirus limnophila corrig. (Hirsch & Muller 1986) Scheuner et al. 2015 [Planctomyces limnophilus Hirsch and Müller 1986]
- Genus Rhodopirellula Schlesner et al. 2004
- Species R. baltica Schlesner et al. 2004 (type sp.)
- Species R. caenicola Yoon et al. 2015
- Species R. rosea Roh et al. 2014
- Species R. rubra Bondoso et al. 2014
- Species R. lusitana Bondoso et al. 2014
- Species "R. europaea" ♠ Frank 2011
- Species "R. islandica" ♠ Kizina et al. 2015
- Species "R. maiorica" ♠ Frank 2011
- Species "R. sallentina" ♠ Frank 2011
- Genus Roseimaritima Bondoso et al. 2015
- Species Roseimaritima ulvae Bondoso et al. 2015
- Species Roseimaritima sediminicola kumar et al. 2020
- Genus Rubinisphaera Scheuner et al. 2015
- Species Rubinisphaera brasiliensis (Schlesner 1990) Scheuner et al. 2015 [Planctomyces brasiliensis Schlesner 1990]
- Genus Rubripirellula Bondoso et al. 2015
- Species Rubripirellula obstinata Bondoso et al. 2015
- Genus Schlesneria Kulichevskaya et al. 2007
- Species Schlesneria paludicola Kulichevskaya et al. 2007
- Genus Singulisphaera Kulichevskaya et al. 2008 emend. Kulichevskaya et al. 2012
- Species "S. mucilaginosa" ♠ Zaicnikova et al. 2011
- Species S. acidiphila Kulichevskaya et al. 2008 (type sp.)
- Species S. rosea Kulichevskaya et al. 2012
- Genus Telmatocola Kulichevskaya et al. 2012
- Species Telmatocola sphagniphila Kulichevskaya et al. 2012
- Genus Thermogutta Slobodkina et al. 2015
- Species T. hypogea Slobodkina et al. 2015
- Species T. terrifontis Slobodkina et al. 2015
- Genus Thermostilla Slobodkina et al. 2015
- Species Thermostilla marina Slobodkina et al. 2015
- Genus Zavarzinella Kulichevskaya et al. 2009
- Species Zavarzinella formosa Kulichevskaya et al. 2009
- Genus "Bythopirellula" ♠ Storesund & Ovreas 2013
- Family Planctomycetaceae Schlesner & Stackebrandt 1987 emend. Scheuner et al. 2014 [Isosphaeraceae Kulichevskaya et al. 2015]
- Class Phycisphaerae Fukunaga et al. 2010
Notes:
♠ Strains found at the National Center for Biotechnology Information (NCBI) but not listed in the List of Prokaryotic names with Standing in Nomenclature (LSPN)
♪ Prokaryotes where no pure (axenic) cultures are isolated or available, i. e. not cultivated or can not be sustained in culture for more than a few serial passages
Ecology
Distribution and abundance
Members of the Planctomycetes are found in a diverse stretch of environments, both geographically and ecologically,[29] and occur in both aquatic and terrestrial habitats.[1] In aquatic environments, they are found in both freshwater ponds and oceans.[29] Furthermore, Planctomycetes were originally believed to exist exclusively in aquatic environments; however, from recent research, it is now known that they are also abundant in soils,[30] as well as hypersaline environments.[31] They are widespread on five continents, including Antarctica and Australia. [30][29]
Planctomycetes are commonly found in aquatic and terrestrial ecosystems. Fluorescence in situ hybridization was used to detect Planctomycetes in various environments and it was determined that Planctomycetes are abundant in Sphagnum bogs. Some were found in the digestive system of marine lifeforms and that some species of Planctomycetes tend to live together with eukaryotes.[4]
Environmental influences on distribution
Planctomycetes account for roughly 11% of prokaryotic communities in marine systems, and their vast distribution demonstrates their ability to inhabit many different environments. They can also adapt to both aerobic and anaerobic conditions. Many factors that can affect their distribution, such as humidity, oxygen levels, and pH. Planctomycete diversity and abundance are strongly associated with relative humidity. The effects of oxygen levels demonstrate the energy needs of the individual. Many species of Planctomycetes are chemoheterotrophic, including G. obscuriglobus. Thermostilla marina, a thermophilic anaerobic species occupying hydrothermal vent regions, can use elemental sulfur to generate sulphide and respire with nitrate. Planctomycetes can also inhabit regions with wide ranges in pH levels, from 4.2 to 11.6.[3]
Ecological impacts and global carbon cycle
Planctomycetes have a significant impact on global biogeochemistry and climate, with their ability to mineralize and break down detritus particles in the water column.[2][14]
Planctomycetes play a considerable role in the global carbon cycle.[1][2][8][32] As both obligate and facultative aerobic chemoheterotrophs, the primary source of carbon used by Planctomycetes is from carbohydrates. Many Planctomycetes have the ability to breakdown extremely complex carbohydrates, therefore making these nutrients available to other organisms. This ability to recycle carbon has been linked to specific C1 metabolism genes observed in many Planctomycetes and are suggested to play a significant role, however this area of research is still poorly understood.
Planctomycetes also display many sulfatase enzymes, which are capable of breaking down sulfated heteropolysaccharides, which are produced by many groups of macroalgae. The breakdown of these sulfated heteropolysaccharides by Planctomycetes are then used as an energy source. It has also been suggested that some Planctomycetes are capable of breaking down carrageenan.[32]
Association with other organisms

Planctomycetes have often been observed in association with many organisms, including, macroalgae, microalgae, marine sponges and plants such as lichen and bryophytes.[3] They have also been observed inhabiting deep sea cold seeps, where they are dominant organisms living on tube worms.[1]
Macroalgae

Planctomycetes are often associated with marine surfaces high in nutrients. They occur as biofilms on algal surfaces in relatively high abundance.[16] Macroalgae such as the kelps Laminaria hyperborea and Ecklonia radiata are suggested to be an important habitat for Planctomycetes.[1][33] Roughly 70% of the bacterial community on Ecklonia radiata were Planctomycetes.[1][5] Almost 150 Planctomycete species have been isolated from the biofilms of macroalgae, and it is suggested that Planctomycete communities associated with macroalgae are mainly independent of changes in geographical distribution. This would suggest a symbiotic relationship.[3]
Kelp forests dominate the rocky coastlines of temperature regions, and provide habitat, shelter, and food for many organisms, including Planctomycetes.[1] Given the considerable role of kelp forests in coastal primary productivity, the association of Planctomycetes with kelp could indicate the significant role of Planctomycetes in coastal habitats.[34] Planctomycetes also play an important role as components of detritus in the water column, also known as marine snow,[1][34] given their ability to attach to surfaces.[35]
Some research has suggested that as the climate continues to warm, the abundance of Planctomycetes associated with macroalgae might increase. The seaweed Caulerpa taxifolia was incubated under higher CO2 conditions, and the abundance of Planctomycetes increased substantially, as much as 10 times in some species.[1]
Mocroalgae and diatom blooms
While macroalgae are well known substrates for Planctomycete communities, the abundance of Planctomycetes has also been known to correlate with blooms of microalgae such as diatoms.[34][1] Blooms of cyanobacteria, diatoms, and dinoflagellates provide nutrients for Planctomycetes, which could explain the association [3]
Marine sponges
Planctomycetes are often associated with the surfaces of marine sponges.[3][35] Planctomycetes interact with sponges either by attachment with a holdfast, or through a symbiotic relationship. A high diversity of Planctomycetes are present as biofilms on sponges. The symbiotic relationship among sponges and Planctomycetes contribute to the health of the sponge, and the sponge often provides suitable habitat and nutrients to the Planctomycetes.[3]
Lichen communities and Sphagnum bogs
Planctomycetes have also been associated with lichen communities and Sphagnum wetlands. Sphagnum wetlands store large amounts of carbon, contributing to the global carbon cycle. Planctomycetes play a considerable role in the degradation of Sphagnum, accounting for roughly 15% of the bacterial community.
Planctomycetes were found to be highly abundant in lichen communities throughout northwestern Siberia. The Planctomycetes associated with these lichen communities also displayed extremely high diversity.[3]
Other bacterial communities
Planctomycetes display associations with other bacterial communities, mainly Alphaproteobacteria, Bacteroidetes, Gemmatimonadetes, and Verrucomicrobia. The growth of many Planctomycetes is often supported by the essential nutrients provided by other bacteria within the community, and some Planctomycetes rely strongly on symbiotic relationships with other bacteria.[3]
Physiology
Endocytosis
Previous studies have suggested that the existence of membrane coat proteins near the intracytoplasmic membrane could be used for an endocytosis-like uptake system, which would be the first instance this function has been found outside of the eukaryotic domain. However, now that the existence of a rigid peptidoglycan cell wall has been confirmed, it seems unlikely for vesicles to be able to pass through this cell wall. Additionally, deletion of one of these membrane coat proteins within P. limnophila found no decrease in macromolecule uptake.[12] Furthermore, with the use of cryo-electron tomography-based three dimensional reconstruction of Planctomycetes has found that what were originally thought to be vesicles being held in the periplasm are actually just folds in the cytoplasmic membrane.[1] Yet it has been demonstrated that Planctomycetes can survive on high molecular weight polysaccharides as their only source of carbon, meaning that Planctomycetes must have the ability to incorporate complex carbon substrates into their cytoplasm. Three hypotheses have been put forth: First, Planctomycetes excrete an enzyme which, outside of the cell wall, degrades the complex substrates into smaller monosaccharides which can more easily be transported through the different membranes. Second, the complex substrates become anchored to the outside of Planctomycetes, which are then able to slowly break down these substrates into oligosaccharides which are able to be transported into the periplasm of Planctomycetes by specialized proteins. The third hypothesis involves the crateriform structures found on the outside of Planctomycete cell walls. These structures have fibers lining their pits that may be able to absorb whole polysaccharides into the periplasm, where they would then be digested.[12]
Osmotic regulation
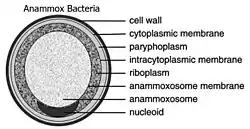
Almost all bacteria have a cytosol following the outer shape of their peptidoglycan cell wall. Eukaryotes are different in that they have their cytosol divided into multiple compartments to create organelles such as a nucleus. Planctomycetes are unique in that they have large invaginations of their cytoplasmic membrane, pulling away from the peptidoglycan cell wall and leaving lots of room for the periplasm. Traditionally, the cytoplasmic membrane has been thought to be responsible for controlling the osmotic pressure of bacterial cells. Yet due to the folds in the cytoplasmic membrane, and the existence of large spaces of periplasm, researchers have suggested that within Planctomycetes their peptidoglycan acts as an osmotic barrier and the periplasm being isotonic to the cytosol.[1]
Anaerobic ammonium oxidation (Anammox)
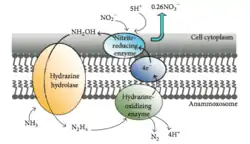
Anammox is the process of oxidizing ammonium where nitrite acts as the electron acceptor. This process creates energy for the organism performing the reaction in the same way humans gain energy from oxidizing glucose.[36] In a marine environment, this ultimately removes nitrogen from the water as N2 gas cannot be used by phytoplankton and will be released into the atmosphere. Up to 67% of dinitrogen gas production in the ocean can be attributed to anammox [37] and about 50% of the nitrogen gas in the atmosphere is thought to be produced from anammox.[38] Planctomycetes are the most dominant phylum of bacteria capable of performing anammox[39] and thus the class of Planctomycetes capable of performing anammox play an important role in the global cycling of nitrogen.
Sterol synthesis
The synthesis of sterols, often observed in eukaryotes and uncommon among bacteria, has been observed very rarely in Planctomycetes.[1][10] The synthesis of sterols such as lanosterol has been observed in G. obscuriglobus. Lanosterol is common in eukaryotes and two other groups of bacteria, both methylotrophic proteobacteria and myxobacteria. The synthesis of sterols observed in G. obscuriglobus is unique within Planctomycetes. Sterol synthesis is suggested to be associated with regulation of membrane fluidity in Planctomycetes,[10] and has been described as essential to the proper growth and reproduction of G. obscuriglobus.[40]
Global and Human Impacts
Climate change
Planctomycetes are widely distributed around the world and are being considered as model organisms in modern microbial research, such as microbial ecology, molecular evolution, and cell biology. They were studied in marine soils that were acidic, pH of 4.2, and hypersaline.
The impacts of research on Planctomycetes and their uses may reach the level of global significance with regards to nutrient cycling processes and assist in furthering understanding for global marine biogeochemistry. However, with Planctomycetes’ growing influences on metabolic processes involving water and air, it may also have a role in interchanges between oceans and atmosphere, potentially affecting climate change.[31]
Biotechnology
There has been recent interest in examining Planctomycetes regarding their potential roles in biotechnology, mainly as a source of bioactive molecules.[3][9] These bioactive molecules are of interest mainly to the pharmaceutical industry. Bioactive compounds are mainly present as secondary metabolites[9], although little is known about planctomycetal secondary metabolites.[41] This is unexpected, as Planctomycetes have several key features that other known producers of bioactive molecules have, such as Myxobacteria.[41] However, there are a number of promising ongoing research that serves as various first steps in including Planctomycetes in small-molecule drug development for humans.
Planctomycetes are worthwhile considerations in challenging the current models for the origin of nucleus, along with other aspects of origin and evolution of the eukaryotic endomembrane system, although extensive studies are still needed.[31]
Planctomycetes as pathogens
Planctomycetes were recently identified as being an opportunistic human pathogen. However, there seems to be a lack of cultured media which is limiting our understanding of how important a role Planctomycetes play in human pathology.[3]
See also
- Anammox, an anaerobic ammonium oxidation process, only found in some of the members of this phylum.
- Global carbon cycle.
- Gemmata obscuriglobus.
- Laminaria hyperborea.
- List of bacterial orders.
References
- Wiegand S, Jogler M, Jogler C (November 2018). "On the maverick Planctomycetes". FEMS Microbiology Reviews. 42 (6): 739–760. doi:10.1093/femsre/fuy029. PMID 30052954.
- Glöckner FO, Kube M, Bauer M, Teeling H, Lombardot T, Ludwig W, et al. (July 2003). "Complete genome sequence of the marine planctomycete Pirellula sp. strain 1". Proceedings of the National Academy of Sciences of the United States of America. 100 (14): 8298–303. Bibcode:2003PNAS..100.8298G. doi:10.1073/pnas.1431443100. PMC 166223. PMID 12835416.
- Kaboré OD, Godreuil S, Drancourt M (2020). "Planctomycetes as Host-Associated Bacteria: A Perspective That Holds Promise for Their Future Isolations, by Mimicking Their Native Environmental Niches in Clinical Microbiology Laboratories". Frontiers in Cellular and Infection Microbiology. 10: 519301. doi:10.3389/fcimb.2020.519301. PMC 7734314. PMID 33330115.
- Wagner, Michael; Horn, Matthias (2006-06-01). "The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance". Current Opinion in Biotechnology. Environmental biotechnology/Energy biotechnology. 17 (3): 241–249. doi:10.1016/j.copbio.2006.05.005. ISSN 0958-1669. PMID 16704931.
- Kohn, Timo; Rast, Patrick; Kallscheuer, Nicolai; Wiegand, Sandra; Boedeker, Christian; Jetten, Mike S. M.; Jeske, Olga; Vollmers, John; Kaster, Anne-Kristin; Rohde, Manfred; Jogler, Mareike (2020). "The Microbiome of Posidonia oceanica Seagrass Leaves Can Be Dominated by Planctomycetes". Frontiers in Microbiology. 11: 1458. doi:10.3389/fmicb.2020.01458. ISSN 1664-302X. PMC 7366357. PMID 32754127.
- Lage OM, Bondoso J, Lobo-da-Cunha A (October 2013). "Insights into the ultrastructural morphology of novel Planctomycetes". Antonie van Leeuwenhoek. 104 (4): 467–76. doi:10.1007/s10482-013-9969-2. PMID 23857394. S2CID 17003623.
- Kumar D, Kumar G, Jagadeeshwari U, Sasikala C, Ramana CV (April 2021). ""Candidatus Laterigemmans baculatus" gen. nov. sp. nov., the first representative of rod shaped planctomycetes with lateral budding in the family Pirellulaceae". Systematic and Applied Microbiology. 44 (2): 126188. doi:10.1016/j.syapm.2021.126188. PMID 33647766.
- Wiegand S, Jogler M, Boedeker C, Pinto D, Vollmers J, Rivas-Marín E, et al. (January 2020). "Cultivation and functional characterization of 79 planctomycetes uncovers their unique biology". Nature Microbiology. 5 (1): 126–140. doi:10.1038/s41564-019-0588-1. PMC 7286433. PMID 31740763.
- Graça AP, Calisto R, Lage OM (2016). "Planctomycetes as Novel Source of Bioactive Molecules". Frontiers in Microbiology. 7: 1241. doi:10.3389/fmicb.2016.01241. PMC 4982196. PMID 27570520.
- Rivas-Marin E, Peeters SH, Claret Fernández L, Jogler C, van Niftrik L, Wiegand S, Devos DP (January 2020). "Non-essentiality of canonical cell division genes in the planctomycete Planctopirus limnophila". Scientific Reports. 10 (1): 66. Bibcode:2020NatSR..10...66R. doi:10.1038/s41598-019-56978-8. PMC 6952346. PMID 31919386.
- Santarella-Mellwig R, Pruggnaller S, Roos N, Mattaj IW, Devos DP (2013-05-21). "Three-dimensional reconstruction of bacteria with a complex endomembrane system". PLOS Biology. 11 (5): e1001565. doi:10.1371/journal.pbio.1001565. PMC 3660258. PMID 23700385.
- Boedeker C, Schüler M, Reintjes G, Jeske O, van Teeseling MC, Jogler M, et al. (April 2017). "Determining the bacterial cell biology of Planctomycetes". Nature Communications. 8 (1): 14853. Bibcode:2017NatCo...814853B. doi:10.1038/ncomms14853. PMC 5394234. PMID 28393831.
- Kallscheuer N, Moreira C, Airs R, Llewellyn CA, Wiegand S, Jogler C, Lage OM (December 2019). "Pink- and orange-pigmented Planctomycetes produce saproxanthin-type carotenoids including a rare C45 carotenoid" (PDF). Environmental Microbiology Reports. 11 (6): 741–748. doi:10.1111/1758-2229.12796. PMID 31600855.
- Viana F, Lage OM, Oliveira R (October 2013). "High ultraviolet C resistance of marine Planctomycetes". Antonie van Leeuwenhoek. 104 (4): 585–95. doi:10.1007/s10482-013-0027-x. hdl:1822/50752. PMID 24052365. S2CID 13153498.
- Kohn T, Heuer A, Jogler M, Vollmers J, Boedeker C, Bunk B, et al. (2016). "Fuerstia marisgermanicae gen. nov., sp. nov., an Unusual Member of the Phylum Planctomycetes from the German Wadden Sea". Frontiers in Microbiology. 7: 2079. doi:10.3389/fmicb.2016.02079. PMC 5177795. PMID 28066393.
- Boersma AS, Kallscheuer N, Wiegand S, Rast P, Peeters SH, Mesman RJ, et al. (December 2020). "Alienimonas californiensis gen. nov. sp. nov., a novel Planctomycete isolated from the kelp forest in Monterey Bay". Antonie van Leeuwenhoek. 113 (12): 1751–1766. doi:10.1007/s10482-019-01367-4. PMID 31802338. S2CID 208641991.
- Kartal, Boran; de Almeida, Naomi M.; Maalcke, Wouter J.; Op den Camp, Huub J.M.; Jetten, Mike S.M.; Keltjens, Jan T. (2013-05-01). "How to make a living from anaerobic ammonium oxidation". FEMS Microbiology Reviews. 37 (3): 428–461. doi:10.1111/1574-6976.12014. ISSN 0168-6445. PMID 23210799.
- Peeters SH, Wiegand S, Kallscheuer N, Jogler M, Heuer A, Jetten MS, et al. (December 2020). "Three marine strains constitute the novel genus and species Crateriforma conspicua in the phylum Planctomycetes". Antonie van Leeuwenhoek. 113 (12): 1797–1809. doi:10.1007/s10482-019-01375-4. PMID 31894495. S2CID 209516837.
- Jogler C, Glöckner FO, Kolter R (August 2011). "Characterization of Planctomyces limnophilus and development of genetic tools for its manipulation establish it as a model species for the phylum Planctomycetes". Applied and Environmental Microbiology. 77 (16): 5826–9. doi:10.1128/AEM.05132-11. PMC 3165242. PMID 21724885.
- Rivas-Marín E, Canosa I, Devos DP (2016). "Evolutionary Cell Biology of Division Mode in the Bacterial Planctomycetes-Verrucomicrobia- Chlamydiae Superphylum". Frontiers in Microbiology. 7: 1964. doi:10.3389/fmicb.2016.01964. PMC 5147048. PMID 28018303.
- Bernander R, Ettema TJ (December 2010). "FtsZ-less cell division in archaea and bacteria". Current Opinion in Microbiology. Growth and development: eukaryotes/prokaryotes. 13 (6): 747–52. doi:10.1016/j.mib.2010.10.005. PMID 21050804.
- Gupta RS, Bhandari V, Naushad HS (2012). "Molecular Signatures for the PVC Clade (Planctomycetes, Verrucomicrobia, Chlamydiae, and Lentisphaerae) of Bacteria Provide Insights into Their Evolutionary Relationships". Frontiers in Microbiology. 3: 327. doi:10.3389/fmicb.2012.00327. PMC 3444138. PMID 23060863.
- Gupta RS (July 2016). "Impact of genomics on the understanding of microbial evolution and classification: the importance of Darwin's views on classification". FEMS Microbiology Reviews. 40 (4): 520–53. doi:10.1093/femsre/fuw011. PMID 27279642.
- Lagkouvardos I, Jehl MA, Rattei T, Horn M (January 2014). "Signature protein of the PVC superphylum". Applied and Environmental Microbiology. 80 (2): 440–5. doi:10.1128/AEM.02655-13. PMC 3911108. PMID 24185849.
- Strous M., Pelletier E., Mangenot S. (06 April 2006). "Deciphering the evolution and metabolism of an anammox bacterium from a community genome". Nature. 440 (7085): 790–794. doi:10.1038/nature04647. PMID 16598256. S2CID 4402553. Check date values in:
|date=(help)CS1 maint: multiple names: authors list (link) - "16S rRNA-based LTP release 123 (full tree)" (PDF). Silva Comprehensive Ribosomal RNA Database. Retrieved 2016-03-20.
- J.P. Euzéby. "Planctomycetes". List of Prokaryotic names with Standing in Nomenclature (LPSN). Retrieved 2016-03-20.
- Sayers; et al. "Planctomycetes". National Center for Biotechnology Information (NCBI) taxonomy database. Retrieved 2016-03-20.
- Fuerst, John (2004). "Planctomycetes: a phylum of emerging interest for microbial evolution and ecology". World Federation for Culture Collections Newsletter. 38. CiteSeerX 10.1.1.538.2883.
- Buckley, Daniel H.; Huangyutitham, Varisa; Nelson, Tyrrell A.; Rumberger, Angelika; Thies, Janice E. (2006-07-01). "Diversity of Planctomycetes in Soil in Relation to Soil History and Environmental Heterogeneity". Applied and Environmental Microbiology. 72 (7): 4522–4531. doi:10.1128/AEM.00149-06. ISSN 0099-2240. PMC 1489350. PMID 16820439.
- Fuerst, J. A. (1995-07-01). "The planctomycetes: emerging models for microbial ecology, evolution and cell biology". Microbiology. 141 (7): 1493–1506. doi:10.1099/13500872-141-7-1493. ISSN 1350-0872. PMID 7551018.
- Woebken D, Teeling H, Wecker P, Dumitriu A, Kostadinov I, Delong EF, et al. (September 2007). "Fosmids of novel marine Planctomycetes from the Namibian and Oregon coast upwelling systems and their cross-comparison with planctomycete genomes". The ISME Journal. 1 (5): 419–35. doi:10.1038/ismej.2007.63. PMID 18043661. S2CID 23859244.
- Bondoso J, Balagué V, Gasol JM, Lage OM (June 2014). "Community composition of the Planctomycetes associated with different macroalgae". FEMS Microbiology Ecology. 88 (3): 445–56. doi:10.1111/1574-6941.12258. PMID 24266389.
- Fuerst JA (October 2005). "Intracellular compartmentation in planctomycetes". Annual Review of Microbiology. 59 (1): 299–328. doi:10.1146/annurev.micro.59.030804.121258. PMID 15910279.
- Izumi H, Sagulenko E, Webb RI, Fuerst JA (October 2013). "Isolation and diversity of planctomycetes from the sponge Niphates sp., seawater, and sediment of Moreton Bay, Australia". Antonie van Leeuwenhoek. 104 (4): 533–46. doi:10.1007/s10482-013-0003-5. PMID 23959164. S2CID 12315225.
- Kartal, Boran (2011). Cultivation, Detection, andEcophysiology of AnaerobicAmmonium-Oxidizing Bacteria. San Diego CA: Academic Press. pp. 89–108. ISBN 978-0-12-381294-0.
- Qian; et al. (2018). "Diversity and distribution of anammox bacteria in water column and sediments of the Eastern Indian Ocean". ScienceDirect. 133: 52–62 – via Elsevier. Explicit use of et al. in:
|last=(help) - Teeseling; et al. (2015). "Anammox Planctomycetes have a peptidoglycan cell wall" (PDF). Nature Communications. 6: 6878. doi:10.1038/ncomms7878. PMC 4432595. PMID 25962786 – via Nature. Explicit use of et al. in:
|last=(help) - Jing; et al. (2015). "A snapshot on spatial and vertical distribution of bacterial communities in the eastern Indian Ocean" (PDF). Acta Oceanologica Sinica. 35 (6): 85–93. doi:10.1007/s13131-016-0871-4. S2CID 89295982 – via SpringerLink. Explicit use of et al. in:
|last=(help) - Rivas-Marin, Elena; Stettner, Sean; Gottshall, Ekaterina Y.; Santana-Molina, Carlos; Helling, Mitch; Basile, Franco; Ward, Naomi L.; Devos, Damien P. (2019-07-02). "Essentiality of sterol synthesis genes in the planctomycete bacterium Gemmata obscuriglobus". Nature Communications. 10 (1): 2916. doi:10.1038/s41467-019-10983-7. ISSN 2041-1723. PMC 6606645. PMID 31266954.
- Jeske, Olga; Jogler, Mareike; Petersen, Jörn; Sikorski, Johannes; Jogler, Christian (2013-10-01). "From genome mining to phenotypic microarrays: Planctomycetes as source for novel bioactive molecules". Antonie van Leeuwenhoek. 104 (4): 551–567. doi:10.1007/s10482-013-0007-1. ISSN 1572-9699. PMID 23982431. S2CID 18752686.
External links
| Wikispecies has information related to Planctomycetes. |