Endornaviridae
Endornaviridae is a family of viruses. Plants, fungi, and oomycetes serve as natural hosts. There are currently 31 species in this family, divided among 2 genera (Alphaendornavirus and Betaendornavirus). Members of Alphaendornavirus infect plants, fungi and the oomycete Phytophthora sp., members of Betaendornavirus infect ascomycete fungi.[1][2][3][4]
| Endornaviridae | |
|---|---|
 | |
| Endornaviridae particle. Shown is the replicative form (dsRNA) of the (+)ssRNA virus. | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Kitrinoviricota |
| Class: | Alsuviricetes |
| Order: | Martellivirales |
| Family: | Endornaviridae |
| Genera | |
|
Alphaendornavirus | |
Taxonomy
The genera recognized by the ICTV (and some species) are as follows:
- Genus Alphaendornavirus; 24 species
- Genus Betaendornavirus; 7 species
- Species Sclerotina sclerotiorum betaendornavirus 1 (type species), with reference strain Sclerotinia sclerotiorum endornavirus 1
- …
Structure

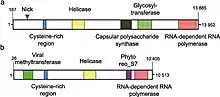
Linear, single-stranded, positive-sense RNA genome of about 14 kb to 17.6 kb. A site specific break (nick) is found in the coding strand about 1 to 2 kb from the 5’ terminus. ViralZone conflicts with ICTV, listing Endornaviridae as dsRNA viruses.[1][2]
As the Endornaviridae genomes don't include a coat protein (CP) gene, they no true virions are associated with members of this family. For Vicia faba endornavirus, the RNA genome has been associated with some pleomorphic cytoplasmic membrane vesicles.[1]
Life cycle
Viral replication is cytoplasmic. The viral replicative form of the Endornaviridae is dsRNA. Replication follows the double-stranded RNA virus replication model. Double-stranded RNA virus transcription is the method of transcription.[2][1]
As the replicative dsRNA form is relatively stable, it can be found in comparatively high quantities in host tissues, and therefore is likely subject of isolations[1] (this is the reason why Endornaviridae', often are classified as dsDNA viruses,[2] in contrast to the official ssRNA(+) ICTV classification).
The virus exits the host cell by cell to cell movement.[1][2]
Plants, fungi, and oomycetes serve as the natural host. Transmission routes are pollen associated.[1][2]
References
- Valverde, RA; Khalifa, ME; Okada, R; Fukuhara, T; Sabanadzovic, S; ICTV Report, Consortium (August 2019). "ICTV Virus Taxonomy Profile: Endornaviridae". The Journal of General Virology. 100 (8): 1204–1205. doi:10.1099/jgv.0.001277. PMID 31184570.
- "Viral Zone". ExPASy. Retrieved 15 June 2015.
- Dolja, Valerian V (2001). "Capsid-Less RNA Viruses". eLS. doi:10.1002/9780470015902.a0023269. ISBN 978-0470016176.
- ICTVdB Management (2006). 00.108.0.01. Endornavirus. In: ICTVdB—The Universal Virus Database, version 4. Büchen-Osmond, C. (Ed), Columbia University, New York, USA.
- NCBI: Oryza sativa endornavirus (isolate Japan) (no rank)
External links
- ICTV: ICTV Report: Endornaviridae
- SIB: Viralzone: Endornaviridae
- NCBI: Endornaviridae (family)
- Syunichi Urayama, Hiromitsu Moriyama, Nanako Aoki, Yukihiro Nakazawa, Ryo Okada, Eri Kiyota, Daisuke Miki, Ko Shimamoto, Toshiyuki Fukuhara: Knock-down of OsDCL2 in rice negatively affects maintenance of the endogenous dsRNA virus, Oryza sativa endornavirus. In: Plant Cell Physiol. 2010 Jan;51(1):58-67. doi:10.1093/pcp/pcp167. PMID 19933266. Epub 2009 Nov 19.